2013 Volume 41 Issue 1 Pages 7-12
2013 Volume 41 Issue 1 Pages 7-12
Rotavirus A causes severe diarrhoea in infants and young children worldwide. The migration pattern (electropherotype) of the double-stranded RNA genome upon polyacrylamide gel electrophoresis has been used to define “strains” in molecular epidemiology. In temperate countries, distinct electropherotypes (strains) appear after the annual, off-seasonal interruption of rotavirus circulation. In Nepal, rotavirus circulated year-round and an uncommon genotype G12P[6] predominated and persisted, providing a unique opportunity to examine whether the same electropherotype (the same strain) persisted or new electropherotypes (new strains) emerged successively under the same G12P[6] predominance. A total of 147 G12P[6] rotaviruses, collected from diarrhoeal children in Nepal between 2007 and 2010, were classified into 15 distinct electropherotypes (strains). Of these, three electropherotypes (strains), LP1, LP24, and LP27, accounted for 10%, 32% and 38% of the G12P[6] rotaviruses, respectively. Each of the three major strains successively appeared, dominated, and disappeared. This study provided new evidence for the hypothesis that rotavirus constantly changes its strains to predominate in the local population even under conditions where a single genotype predominates and persists. Such dynamic strain replacement, the constant takeover of one predominant strain by another, fitter strain, is probably gives a competitive edge to the survival of rotavirus in nature.
Rotavirus A, a member of genus Rotavirus within family Reoviridae [1], causes severe acute diarrhoea in infants and young children worldwide [2] and is the cause of an estimated 453,000 deaths among children less than five years of age occurring each year mostly in developing countries [3]. The genome of rotavirus consists of 11 segments of double-stranded RNA encased within a non-enveloped, triple-layered capsid [1]. Upon polyacrylamide gel electrophoresis (PAGE), the migration pattern of the genomic RNA segments, termed electropherotype, shows considerable variation, and electropherotyping has been used to identify an individual strain in molecular epidemiological studies [1]. Individual strains of rotaviruses are also described by the combination of the genotypes of the two capsid proteins VP4 and VP7 encoded, respectively, by genome segment 4 and genome segment 7, 8, or 9 depending on the strain [1]. These two proteins are the target of neutralizing antibodies and the genotype (serotype) defined by VP4 is referred to as the P type since VP4 is protease-sensitive whereas that defined by VP7 is referred to as the G type since VP7 is a glycoprotein [1]. While more than two dozen alleles have been reported for both G and P types, common human rotaviruses are known to carry genotype combinations of G1P[8], G2P[4], G3P[8], G4P[8] and G9P[8], together accounting for >80% of human rotavirus genotypes [4–6].
It has been shown that multiple electropherotypes are present within a genotype but that the rotaviruses with the same electropherotype always have the same G and P types [7, 8]. In a 10-year study carried out in Akita, Japan between 1987 and 1996 [9], it was found that one predominant electropheerotype (occasionally two electropheerotypes) emerged every rotavirus season, rarely persisting to the following season, and that the predominant electropheerotype changed from season to season. In another study carried out in Melbourne, Australia between 1973 and 1979 [10], there was a limited diversity of electropherotype of rotaviruses was observed in neonates, one of which persisted for four years, whereas much greater diversity and variability with a shorter circulation period for each electropherotype were observed in the electropheerotypes of rotaviruses from children and adults.
However, these observations were made in areas where the circulation of rotavirus is seasonal with a period of no rotavirus detection between two seasons [7, 9, 10]. In contrast, the circulation of rotavirus was year-round in Nepal where we carried out a hospital-based surveillance for seven years. In addition, there was continued predominance of rotaviruses possessing G12P[6] with an average detection rate of 30% since late 2003 [11–14]. These unique features of the epidemiology of rotavirus in Nepal provided, for the first time, an opportunity to determine whether the same electropherotype persisted or whether new electropherotypes appeared one after another within the single predominant genotype combination of G12P[6] in a defined local population of children less than five years of age.
A total of 539 rotavirus-positive stool specimens were collected from children admitted to Kanti Children’s Hospital in Kathmandu, Nepal for the treatment of acute gastroenteritis between November 2007 and February 2010. A portion of the results was published previously [13, 14], and the entire surveillance report will be published elsewhere.
RNA extraction and PAGEGenomic double-stranded RNAs were extracted with a QIAamp Viral RNA Mini Kit (QIAGEN Sciences, Germantown, MD, USA) from 10% (w/v) stool suspension of rotavirus-positive stool specimens initially identified with a commercial enzyme-linked immunosorbent assay kit (Rotaclone; Meridian Diagnostics, Cincinnati, OH, USA) as described previously [13]. For the purpose of assigning an electropherotype for each rotavirus specimen, the genomic RNAs were separated by electrophoresis on a 10% polyacrylamide gel (0.75 mm in thickness) with a 4% stacking gel in the Laemmli buffer system using an SE600 Ruby electrophoresis apparatus (GE Healthcare Bio-Science Corp. Picataway, NJ, USA ). When the result by comparison on a 10% gel was ambiguous, a 7.5% or 12.5% gel was also used as described previously [8]. After electrophoresis for 16 h at a constant current of 8 mA per gel, the gels were stained with 0.19 %(w/v) silver nitrate for 1 h and treated with 3 %(w/v) sodium hydroxide and 0.5 %(v/v) formaldehyde until the bands became visible. Genomic RNAs from strains Wa (G1P[8]) and KUN (G2P[4]) were included in each gel as reference for long and short RNA pattern, respectively. Samples on different gels with a similar RNA migration profile were run together on the same gel for further comparison. Co-electrophoresis was carried out for samples with a similar RNA profile yet with different G or P genotypes. A rotavirus strain was defined by the distinct RNA migration pattern on PAGE.
Identification of stool specimens containing G12P[6] rotavirusesGenotyping of rotavirus strains was performed by reverse-transcription (RT)-PCR according to the method described by Gouvea et al. [15] and Gunasena et al. [16] for G and P types, respectively. Of 539 rotavirus-positive stool specimens, 185 (34%) were identified as G12P[6]. Details of genotype distributions will be published elsewhere.
The number of G12P[6] stool rotavirus specimens to which electropherotypes were assigned.Of 185 G12P[6] rotaviruses, the presence of rotavirus-specific RNA was identified in 172 (93%) specimens, and 147 showed RNA patterns clear enough to assign electropherotype. Thus, electropherotypes were assigned to 79% of the G12P[6] rotaviruses.
Among a total of 147 G12P[6] rotavirus specimens in which a sufficient amount of genomic RNA was present, 15 electropherotypes were distinguished by side-by-side comparison or by co-electrrophoresis, indicating that there were 15 distinct G12P[6] rotavirus strains circulating over the 28-month period between October 2007 and February 2010 (Fig. 1). Of those 15 electropherotypes, three major electropherotypes comprised 80% of the G12P[6] rotaviruses, with LP1, LP24 and LP27 accounting for 10%, 32% and 38%, respectively. The other 12 electropherotypes accounted for the remaining 20%.
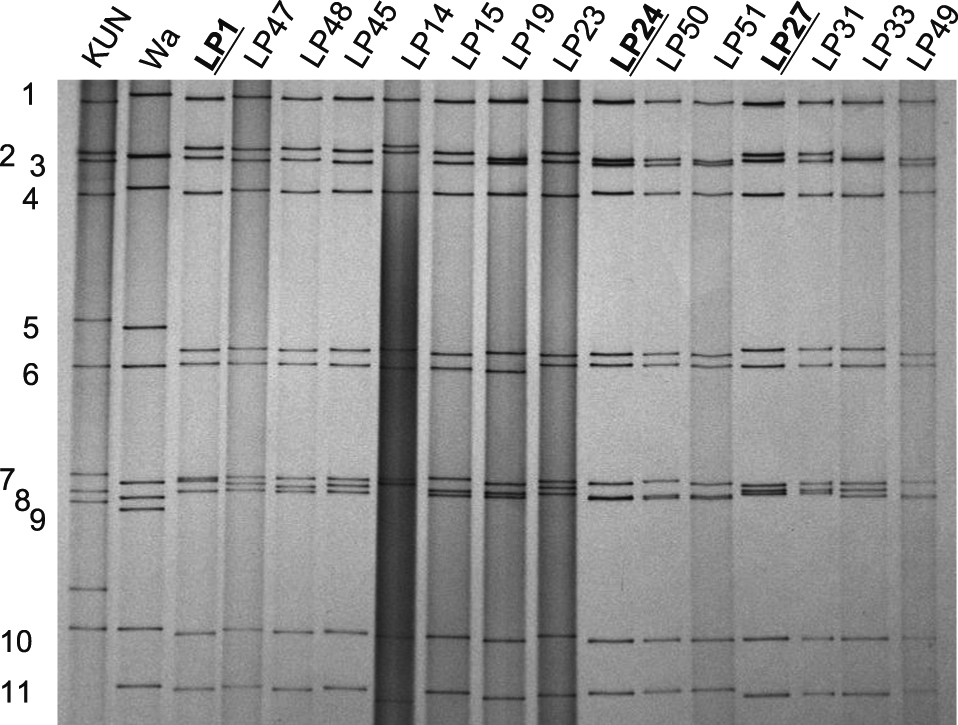
The 15 distinct electropherotypes (strains) of the genomic RNA segments from the G12 P[6] rotaviruses detected from children with severe diarrhoea in Kathmandu, Nepal between October 2007 and February 2010. KUN and Wa are reference strains representing a rotavirus with a short RNA pattern (faster moving 10th and 11th genome segments) and a rotavirus with a long RNA pattern (slower moving 10th and 11th genome segments), respectively. The name of an electropherotype (hence the name of a strain) is shown on the top of each lane on the gel. Approximate positions of the genome segments of Wa are indicated to the left of the gel. A 10% polyacrylamide gel was stained with silver nitrate.
When the three predominant electropherotypes were compared side by side on gels with three different acrylamide concentrations of 7.5%, 10% and 12.5% and by co-electrophoresis on a 12.5% gel, differences were observed between LP1 and LP24 in the migration rate of six cognate genome segments, between LP24 and LP27 in the migration rate of seven segments, and between LP1 and LP27 in the migration rate of nine segments (Table 1). There were only two genome segments (i.e., segments 1 and 10) that migrated at the same positions across the three predominant electropherotypes (Table 1). Thus, the three major electropherotypes differed among each other in more than half of their genome segments.
| Genome Segment | LP 1 (n = 15) | LP24 (n = 47) | LP27 (n = 56) |
|---|---|---|---|
| 1 | a | a | a |
| 2 | a | b (fastest*) | c (faster) |
| 3 | a | b (faster**) | c (slower**) |
| 4 | a | b (slower) | b (slower) |
| 5 | a | a | c (slower) |
| 6 | a | b (slower) | c (faster) |
| 7 | a | b (faster) | b (faster) |
| 8 | a | b (fastest) | c (faster) |
| 9 | a | a | c (slower) |
| 10 | a | a | a |
| 11 | a | a | c (faster) |
In each genome segment, the position on the gel at which the RNA segment of LP1 migrated is designated as “a”. Position “b” is designated when any change in the migrating position on the gel from “a” is observed. Similarly, position “c” is designated when any further change from positions “a” and “b” is observed.
* indicated as “fastest” when the migration of a particular genome segment is the fastest among the cognate genome segments of the three electropheotypes.
** indicated as “faster” or “slower” when the migration of a particular genome segment is either faster or slower than the cognate segment of the preceding strain.
When we examined the monthly distribution of the three predominant electropherotypes along the time line of the 28-month period (2 years and 4 months), LP1 made its first appearance in November 2007 and dominated until April 2008, accounting for 62% of the G12P[6] rotaviruses detected during its predominant period (Fig. 2). A few months after LP1 disappeared, LP24 made its first appearance in October 2008 and dominated until March 2009, accounting for 79% of the G12P[6] rotaviruses detected during its predominant period. By contrast, LP27 made its first appearance in March 2009 when LP24 was still predominant, subsequently increased its prevalence to 60% in the following nine months, and finally reached to 100% in the last two months of January and February 2010, with an overall prevalence of 67% (Fig. 2). Thus, the prevalence exceeded 60% in the period when each of the three electropherotypes was predominant.
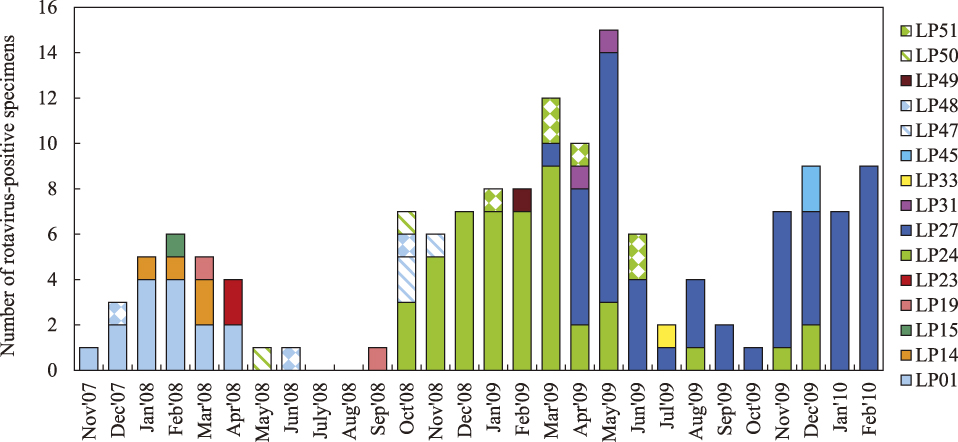
Monthly distribution of the 15 distinct electropherotypes (strains) of rotaviruses possessing G12P[6] detected from children with severe diarrhoea in Kathmandu, Nepal between October 2007 and February 2010.
Strain is an ambiguous term in microbiology [17] but serves as a fundamental unit in the molecular epidemiology of rotaviruses. Studies defining a strain of rotavirus as a group of rotaviruses that share an identical electropherotype provided an important insight into how rotaviruses maintained themselves in nature [7, 9]. Taking advantage of year-round circulation of rotaviruses and the continued predominance of a single genotype of G12P[6] in Kathmandu, Nepal over a 28-month period, the present study showed that the predominance of G12P[6] rotaviruses was maintained such that one preceding strain was replaced by another strain after persisting for several months. An obvious implication from this successive replacement of predominant strains within a single predominant genotype is that replacement probably resulted from the operation of natural selection on the pool of co-circulating strains, that is, a process of selecting a strain fitter than its precursor in its capacity to spread among the susceptible population. At almost every time point, strains with less-frequently detected electropherotypes probably represented left-out strains as well as the pool of strains from which future predominant strains can arise.
To the best of our knowledge, this study is the first to describe strain replacement, at the level of strain, as a mechanism to explain the continued dominance of any single genotype in a defined population. However, a few preceding studies discussed the occurrence of lineage replacement in the gene for VP7, the major neutralization protein defining G type, was discussed to explain the continued dominance of rotavirus with a single genotype. For example, Arista et al. [18] sequenced the VP7 genes of the predominant strains selected on the basis of their electropherotypes from a rotavirus collection over a 19-year period in Palermo, Italy. There were periods of the continued predominance of G1P[8] rotaviruses each spanning over a few years. Arista et al. [18] analyzed how lineages of the G1 VP7 gene changed during each period of the G1P[8] predominance and discovered lineage switching: various lineages and sublineages appearing, disappearing, or co-circulating in an alternate fashion. On the basis of these observations, they postulated a hypothesis that the introduction of a G1 lineage new to the local population was responsible for the continuous circulation of G1 rotaviruses in that population probably under the influence of immune-pressure mechanisms. As another example, Doan et al. [19] performed a comprehensive molecular phylogenetic analysis of the global collection of the G2 VP7 genes deposited over a period of 34 years in the DNA databases, and hypothesized that the G2 VP7 genes evolved in a dynamic fashion, new lineages emerging within the pool of the previous lineages and subsequently dominating. Taken together, the results strongly suggest that rotavirus evolves by a mechanism of successive replacement not only at the level of the VP7 gene but at the level of the strain as a whole.
There is, however, one major limitation to this study. While electropherotype is concerned with all genome segments and has served as the best practical fingerprinting tool in tracking the chain of rotavirus transmission, it fails to provide information about antigenic properties including G and P genotypes of the strain in question. This is the major disadvantage of electropherotyping because strain replacement very likely results from immune selection on the neutralization proteins. Thus, strains with two different elctropherotypes may carry the same VP7 or VP4 gene, the genes coding for the virus neutralization antigens. Conversely, the strains with the same electropherotype may carry the VP7 or VP4 gene that differs so slightly in their nucleotide sequences that no visible migration difference can be observed on PAGE. However, such changes may affect critical amino acid residues for virus neutralization. Thus, while some strains, LP27 for example, were interpreted to have selective advantages over other concurrently circulating strains, it was impossible to ascribe selective advantages to any genome segments let alone to any amino acid residues. Despite this disadvantage, only electropherotyping, and not phylogenetic analysis at the single genome segment level, allows the classification of a large number of field rotavirus specimens into strains based on the overall genomic RNA constellation.
In conclusion, this study provided new evidence, at the level of strain, to support the hypothesis that rotavirus constantly changes its strains to predominate in the local population even under conditions in which a single genotype dominated and persisted. Such dynamic strain replacement, the constant takeover of one predominant strain by another fitter strain, probably gives a competitive edge to the survival of rotavirus in nature.
There is no conflict of interest for any author to declare regarding this study.
Punita Gauchan is a Ph.D. student supported by a scholarship from the Ministry of Education, Culture, Sports, Science and Technology, Japan. N. A. Cunliffe participated in this study according to the Agreement on Academic Partnership between The University of Liverpool and Nagasaki University.
This study was supported by a grant-in-aid for overseas scientific research from Japan Society for the Promotion of Science awarded to Osamu Nakagomi (Nos. 16406016 and 20406013). This study was in part supported by grants-in-aid for scientific research from the Ministry of Health, Welfare and Labour, Japan. This study was also supported by the Global Center of Excellence Program on Integrated Global Control Strategy for Tropical and Emerging Infectious Diseases from the Ministry of Education, Culture, Sports, Science and Technology, Japan.